Five protein-design questions that still challenge AI
You can also search for this author in PubMed Google Scholar
Alena Khmelinskaia wants designing bespoke proteins to be as simple as ordering a meal. Picture a vending machine, she says, which any researcher could use to specify their desired protein’s function, size, location, partners and other characteristics. “Ideally, you would get the perfect design that can accomplish all these things together,” says Khmelinskaia, a biophysical chemist at Ludwig Maximilian University in Munich, Germany.
For the moment, that is just a dream. But advances in computational protein design and machine learning are bringing it closer to reality than ever.
Until a few years ago, researchers altered proteins by cloning them into bacteria or yeast, and coaxing the microorganisms to mutate until they produced the desired product. Scientists could also design a protein manually by deliberately altering its amino-acid sequence, but that’s a laborious process that could cause it to fold incorrectly or prevent the cell from producing it at all.
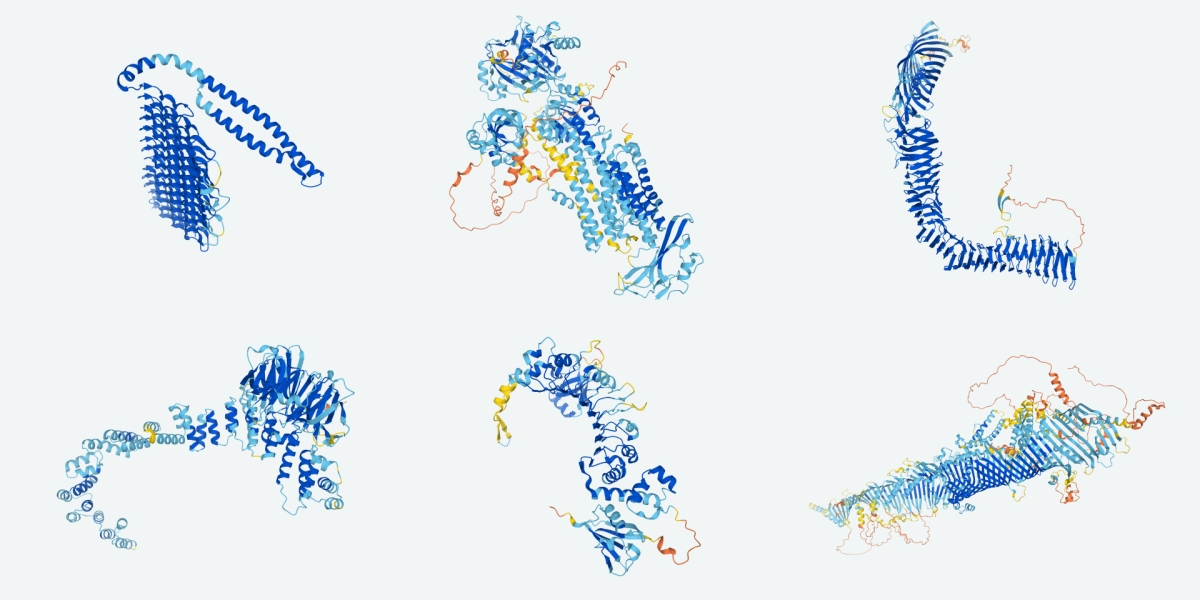
Machine-learning algorithms have changed the game entirely. Researchers can generate new protein structures on their laptops using tools driven by artificial intelligence (AI), such as RFdiffusion and Chroma, which were trained on hundreds of thousands of structures in the Protein Data Bank (PDB). They can identify a sequence to match that structure using algorithms such as ProteinMPNN. RoseTTAFold and AlphaFold, which calculate structures from a sequence, can predict whether the new protein is likely fold correctly. Only then do researchers need to synthesize the physical protein and test whether it works as predicted.