In vivo CRISPR base editing of PCSK9 durably lowers cholesterol in primates
Nature volume 593, pages 429–434 (2021 )Cite this article
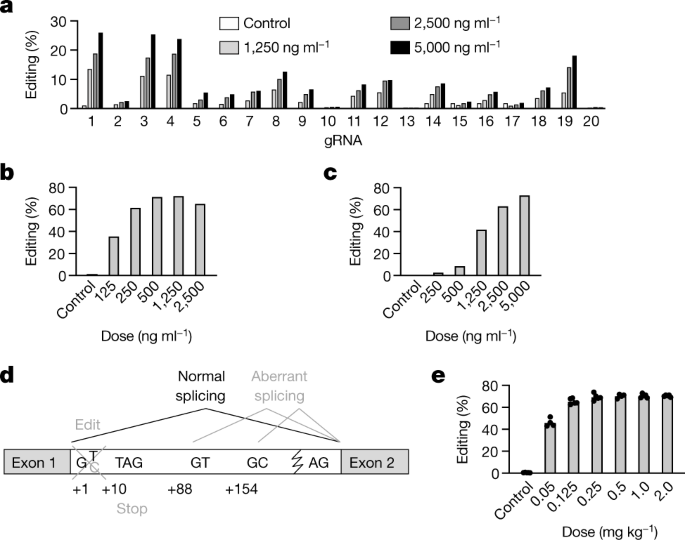
Gene-editing technologies, which include the CRISPR–Cas nucleases1,2,3 and CRISPR base editors4,5, have the potential to permanently modify disease-causing genes in patients6. The demonstration of durable editing in target organs of nonhuman primates is a key step before in vivo administration of gene editors to patients in clinical trials. Here we demonstrate that CRISPR base editors that are delivered in vivo using lipid nanoparticles can efficiently and precisely modify disease-related genes in living cynomolgus monkeys (Macaca fascicularis). We observed a near-complete knockdown of PCSK9 in the liver after a single infusion of lipid nanoparticles, with concomitant reductions in blood levels of PCSK9 and low-density lipoprotein cholesterol of approximately 90% and about 60%, respectively; all of these changes remained stable for at least 8 months after a single-dose treatment. In addition to supporting a ‘once-and-done’ approach to the reduction of low-density lipoprotein cholesterol and the treatment of atherosclerotic cardiovascular disease (the leading cause of death worldwide7), our results provide a proof-of-concept for how CRISPR base editors can be productively applied to make precise single-nucleotide changes in therapeutic target genes in the liver, and potentially in other organs.
DNA and RNA sequencing data that support the findings of this study have been deposited in the NCBI Sequence Read Archive with the accession code PRJNA716270. All other data supporting the findings of this study (Figs. 1–4, Extended Data Figs. 1–9) are available within the Article and its Supplementary Information. The GRCh38 reference human genome (ftp.ncbi.nlm.nih.gov/genomes/all/GCA/000/001/405/GCA_000001405.15_GRCh38/seqs_for_alignment_pipelines.ucsc_ids/GCA_000001405.15_GRCh38_no_alt_analysis_set.fna.gz, ftp://ftp.ensembl.org/pub/release-98/fasta/homo_sapiens/dna/Homo_sapiens.GRCh38.dna.chromosome.{1-22,X,Y,MT}.fa and ftp://ftp.ensembl.org/pub/release-98/fasta/homo_sapiens/dna/Homo_sapiens.GRCh38.dna.nonchromosomal.fa) and Gencode v.34 (ftp://ftp.ebi.ac.uk/pub/databases/gencode/Gencode_human/release_34/gencode.v34.primary_assembly.annotation.gtf.gz) and Ensembl v.98 (ftp://ftp.ensembl.org/pub/release-98/gtf/homo_sapiens/Homo_sapiens.GRCh38.98.gtf.gz) annotations were used. The macFas5 cynomolgus monkey reference genome (ftp://ftp.ensembl.org/pub/release-98/fasta/macaca_fascicularis/dna/Macaca_fascicularis.Macaca_fascicularis_5.0.dna.chromosome.{1-20,X,MT}.fa.gz and ftp://ftp.ensembl.org/pub/release-98/fasta/macaca_fascicularis/dna/Macaca_fascicularis.Macaca_fascicularis_5.0.dna.nonchromosomal.fa.gz) was used. Source data are provided with this paper.
Leave a Comment
Related Posts