Temporal dynamics of the multi-omic response to endurance exercise training
Nature volume 629, pages 174–183 (2024 )Cite this article
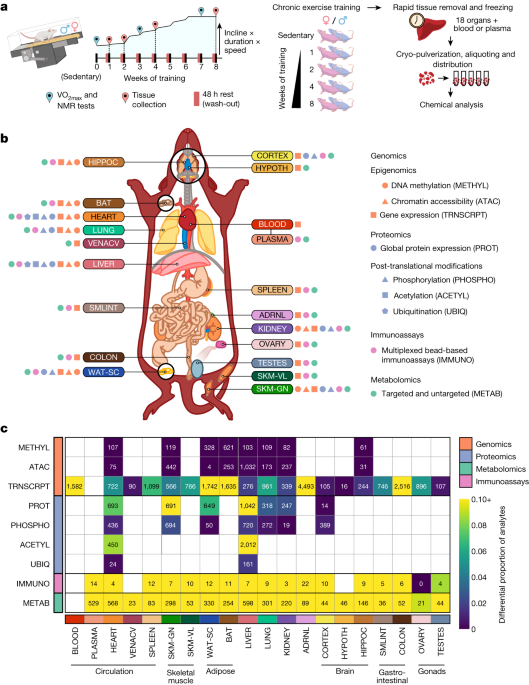
Regular exercise promotes whole-body health and prevents disease, but the underlying molecular mechanisms are incompletely understood1,2,3. Here, the Molecular Transducers of Physical Activity Consortium4 profiled the temporal transcriptome, proteome, metabolome, lipidome, phosphoproteome, acetylproteome, ubiquitylproteome, epigenome and immunome in whole blood, plasma and 18 solid tissues in male and female Rattus norvegicus over eight weeks of endurance exercise training. The resulting data compendium encompasses 9,466 assays across 19 tissues, 25 molecular platforms and 4 training time points. Thousands of shared and tissue-specific molecular alterations were identified, with sex differences found in multiple tissues. Temporal multi-omic and multi-tissue analyses revealed expansive biological insights into the adaptive responses to endurance training, including widespread regulation of immune, metabolic, stress response and mitochondrial pathways. Many changes were relevant to human health, including non-alcoholic fatty liver disease, inflammatory bowel disease, cardiovascular health and tissue injury and recovery. The data and analyses presented in this study will serve as valuable resources for understanding and exploring the multi-tissue molecular effects of endurance training and are provided in a public repository (https://motrpac-data.org/).
Regular exercise provides wide-ranging health benefits, including reduced risks of all-cause mortality1,5, cardiometabolic and neurological diseases, cancer and other pathologies2,6,7. Exercise affects nearly all organ systems in either improving health or reducing disease risk2,3,6,7, with beneficial effects resulting from cellular and molecular adaptations within and across many tissues and organ systems3. Various ‘omic’ platforms (‘omes’) including transcriptomics, epigenomics, proteomics and metabolomics, have been used to study these events. However, work to date typically covers one or two omes at a single time point, is biased towards one sex, and often focuses on a single tissue, most often skeletal muscle, heart or blood8,9,10,11,12, with few studies considering other tissues13. Accordingly, a comprehensive, organism-wide, multi-omic map of the effects of exercise is needed to understand the molecular underpinnings of exercise training-induced adaptations. To address this need, the Molecular Transducers of Physical Activity Consortium (MoTrPAC) was established with the goal of building a molecular map of the exercise response across a broad range of tissues in animal models and in skeletal muscle, adipose and blood in humans4. Here we present the first whole-organism molecular map of the temporal effects of endurance exercise training in male and female rats and provide multiple insights enabled by this MoTrPAC multi-omic data resource.















/cdn.vox-cdn.com/uploads/chorus_asset/file/24748328/236706_Mac_Pro_AKrales_0094.jpg)