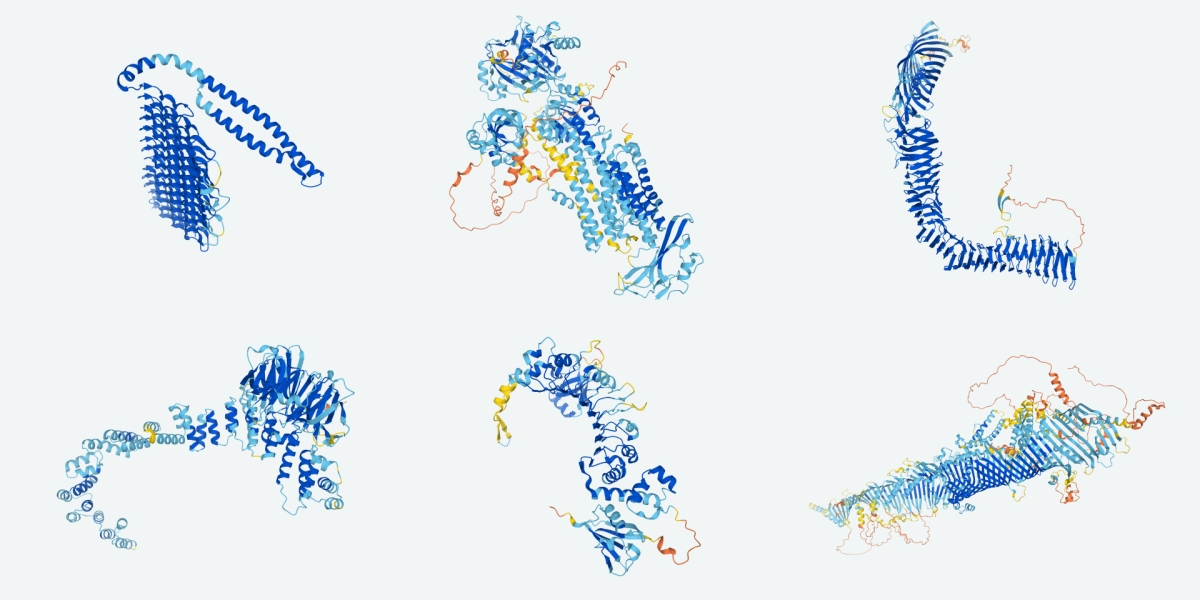
Accurate prediction of protein structures and interactions using a three-track neural network
DeepMind presented remarkably accurate predictions at the recent CASP14 protein structure prediction assessment conference. We explored network architectures incorporating related ideas and obtained the best performance with a three-track network in which information at the 1D sequence level, the 2D distance map level, and the 3D coordinate level is successively transformed and integrated. The three-track network produces structure predictions with accuracies approaching those of DeepMind in CASP14, enables the rapid solution of challenging X-ray crystallography and cryo-EM structure modeling problems, and provides insights into the functions of proteins of currently unknown structure. The network also enables rapid generation of accurate protein-protein complex models from sequence information alone, short circuiting traditional approaches which require modeling of individual subunits followed by docking. We make the method available to the scientific community to speed biological research.
The prediction of protein structure from amino acid sequence information alone has been a longstanding challenge. The bi-annual Critical Assessment of Structure (CASP) meetings have demonstrated that deep learning methods such as AlphaFold (1, 2) and trRosetta (3), that extract information from the large database of known protein structures in the PDB, outperform more traditional approaches that explicitly model the folding process. The outstanding performance of DeepMind’s AlphaFold2 in the recent CASP14 meeting (https://predictioncenter.org/casp14/zscores_final.cgi) left the scientific community eager to learn details beyond the overall framework presented and raised the question of whether such accuracy could be achieved outside of a world-leading deep learning company. As described at the CASP14 conference, the AlphaFold2 methodological advances included 1) starting from multiple sequence alignments (MSAs) rather than from more processed features such as inverse covariance matrices derived from MSAs, 2) replacement of 2D convolution with an attention mechanism that better represents interactions between residues distant along the sequence, 3) use of a two-track network architecture in which information at the 1D sequence level and the 2D distance map level is iteratively transformed and passed back and forth, 4) use of an SE(3)-equivariant Transformer network to directly refine atomic coordinates (rather than 2D distance maps as in previous approaches) generated from the two-track network, and 5) end-to-end learning in which all network parameters are optimized by backpropagation from the final generated 3D coordinates through all network layers back to the input sequence.