CASP14: what Google DeepMind’s AlphaFold 2 really achieved, and what it means for protein folding, biology and bioinformatics
Disclaimer: this post is an opinion piece based on the experience and opinions derived from attending the CASP14 conference as a doctoral student researching protein modelling. When provided, quotes have been extracted from my notes of the event, and while I hope to have captured them as accurately as possible, I cannot guarantee that they are a word-by-word facsimile of what the individuals said. Neither the Oxford Protein Informatics Group nor I accept any responsibility for the content of this post.
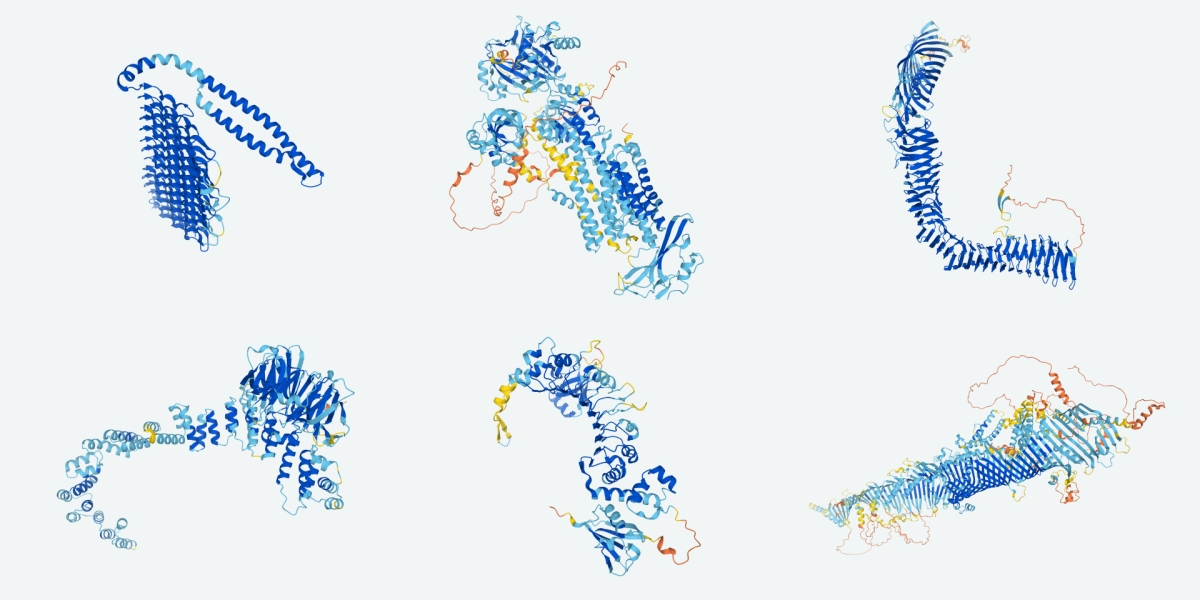
You might have heard it from the scientific or regular press, perhaps even from DeepMind’s own blog. Google ‘s AlphaFold 2 indisputably won the 14th Critical Assessment of Structural Prediction competition, a biannual blind test where computational biologists try to predict the structure of several proteins whose structure has been determined experimentally — yet not publicly released. Their results are so incredibly accurate that many have hailed this code as the solution to the long-standing protein structure prediction problem.
Protein structure is at the core of biochemistry, and has profound implications for medicine and technology. Establishing the structure of a protein is a bottleneck in structure-based drug discovery, and accurate structure prediction is expected to improve the productivity of pharmaceutical research pipelines (although it is only one factor, and we will need to get other things right before truly revolutionary changes happen — check Derek Lowe’s posts here and here). Structural information of proteins is also essential in biology, where it helps to elucidate function — many key papers in biochemistry derive insight from experimental advances in structure determination.