More Protein Folding Progress – What’s It Mean?
Derek Lowe's commentary on drug discovery and the pharma industry. An editorially independent blog from the publishers of Science Translational Medicine. All content is Derek’s own, and he does not in any way speak for his employer.
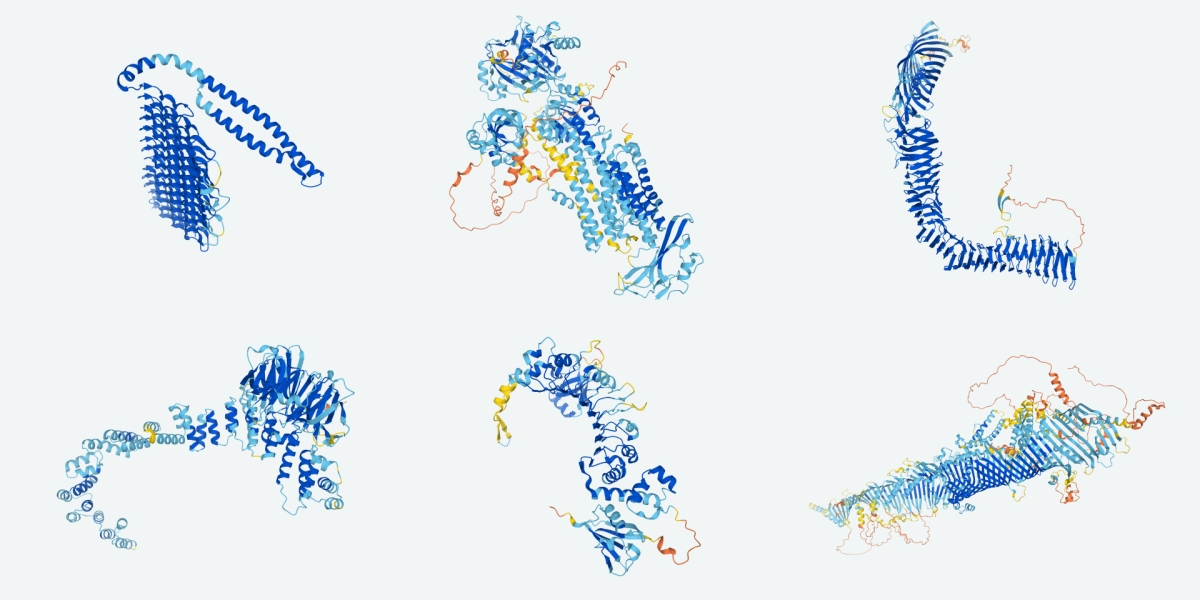
I last wrote about Deepmind’s efforts to predict protein folding and structure here, with their AlphaFold software. AlphaFold really performed very strongly in the 2020 protein folding challenge, and that got a lot of attention. Well, they’ve recently published a great deal of detail on how they did this, released their source code, and they’ve announced that they’re going to be releasing their computed structures of 350,000 proteins, to be followed in the coming months by up to 100 million more. Here’s the database. And a group at the University of Washington has just published on their own similar approach (RosettaFold) and has also made this code freely available.
So it’s clear that the world of computational protein structure prediction is in a very different place than it was a couple of years ago. But I’m already on record as (1) cheering this sort of thing on while (2) saying that it doesn’t make as much difference to drug discovery as many stories and press releases have had it. Do the latest developments change my mind? What does this all mean? I’ve been talking with colleagues and seeing comments from people in the field, so here’s my attempt.















/https%3A%2F%2Ftf-cmsv2-smithsonianmag-media.s3.amazonaws.com%2Ffiler_public%2F6f%2F64%2F6f64a534-be29-44a3-a883-0c19b5e99984%2Fab1_-_branched_bamboo_coral_3689_7-25-09_jsl_0589.jpg)