Delocalization-Induced Molecular Equality
Molecular representation underpins all of cheminformatics and, arguably, chemistry itself. Getting molecular representation right is hard because it supports so many applications, and because all forms of molecular representation have limitations. When a molecular representation system finds its way into applications for which it's underqualified, the result can range from annoying to catastrophic. Today's article describes a phenomenon responsible for many mispairings of representation with application.
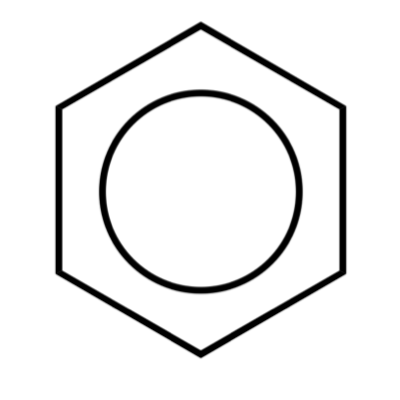
Although molecular structure can be misrepresented in many ways, an especially rich source of errors is bonding. Cheminformatics draws most of its ideas about bonding from the valence bond (VB) model. This model views a bond as the interaction between two atoms and 2n electrons, where n is a positive integer. Cheminformatics adapts this idea by representing molecules as graphs onto which atom/bond electron counts are overlaid. As a result, the formal bond order computed across any edge in a VB graph will be a positive integer.
George Box famously observed that "All models are wrong, but some are useful." It doesn't take much effort to understand that the VB model is wrong. However, it can take a lot of effort to understand when and why the VB model isn't useful. I'll focus on the VB model, but it's important to keep in mind that any molecular representation system will by necessity be limited and therefore wrong in the right context.
Leave a Comment
Related Posts

Cis-urocanic acid, a sunlight-induced immunosuppressive factor, activates immune suppression via the 5-HT2A receptor
Comment














/cdn.vox-cdn.com/uploads/chorus_asset/file/25408886/post_logo.png)


