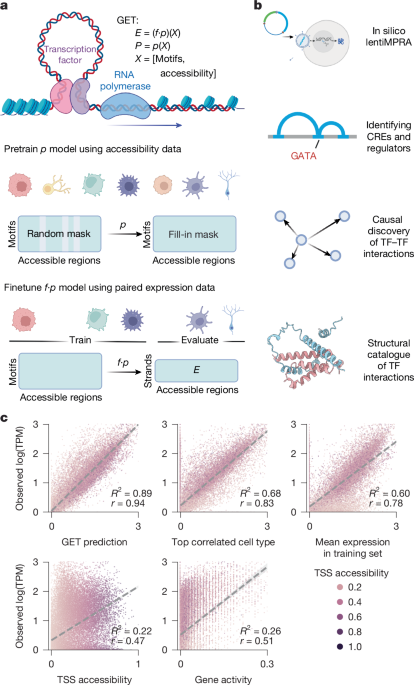
A foundation model of transcription across human cell types
Transcriptional regulation, which involves a complex interplay between regulatory sequences and proteins, directs all biological processes. Computational models of transcription lack generalizability to accurately extrapolate to unseen cell types and conditions. Here we introduce GET (general expression transformer), an interpretable foundation model designed to uncover regulatory grammars across 213 human fetal and adult cell types1,2. Relying exclusively on chromatin accessibility data and sequence information, GET achieves experimental-level accuracy in predicting gene expression even in previously unseen cell types3. GET also shows remarkable adaptability across new sequencing platforms and assays, enabling regulatory inference across a broad range of cell types and conditions, and uncovers universal and cell-type-specific transcription factor interaction networks. We evaluated its performance in prediction of regulatory activity, inference of regulatory elements and regulators, and identification of physical interactions between transcription factors and found that it outperforms current models4 in predicting lentivirus-based massively parallel reporter assay readout5,6. In fetal erythroblasts7, we identified distal (greater than 1 Mbp) regulatory regions that were missed by previous models, and, in B cells, we identified a lymphocyte-specific transcription factor–transcription factor interaction that explains the functional significance of a leukaemia risk predisposing germline mutation8,9,10. In sum, we provide a generalizable and accurate model for transcription together with catalogues of gene regulation and transcription factor interactions, all with cell type specificity.
Transcriptional regulation constitutes a critical yet largely unresolved domain that underpins diverse biological processes, including those associated with human genetic diseases and cancers11. Transcriptional changes are orchestrated by a conserved regulatory machinery, including transcription factors (TFs) that bind to regulatory sequences; coactivators, mediators, and core transcriptional factors; and RNA polymerase II (PolII)12. Although different cell types may possess different subsets of regulatory regions, the biochemistry of protein–protein and protein–DNA interactions remains largely the same across cell types when epigenetic conditions are fixed. Clustering of known TF binding site motifs13 demonstrates significant homology in TF DNA-binding domains, further reducing the combinatorial variability of regulatory interactions. However, our understanding of transcription regulation is often limited to specific cell types, and it is not clear how the combinatorial interaction of different TFs determines the diversity of expression profiles observed across cell types. As an example, previous expression prediction methods such as Expecto14, Basenji2 (ref. 15) and Enformer4 are designed to make predictions on the training cell types after they have been fine-tuned, hindering the generalizability and utility of these models.