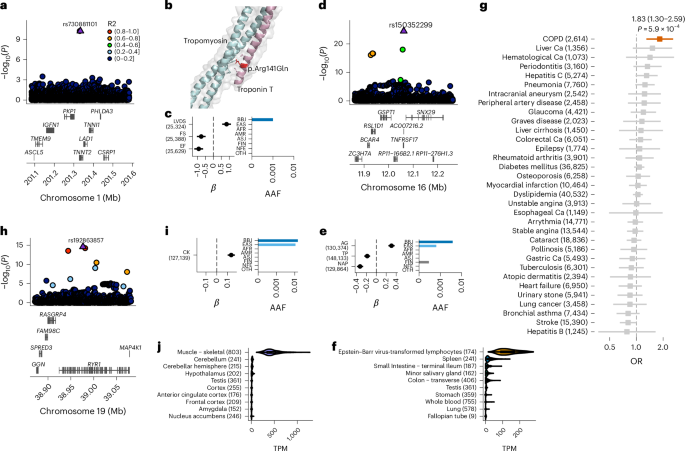
Population-specific putative causal variants shape quantitative traits
Human genetic variants are associated with many traits through largely unknown mechanisms. Here, combining approximately 260,000 Japanese study participants, a Japanese-specific genotype reference panel and statistical fine-mapping, we identified 4,423 significant loci across 63 quantitative traits, among which 601 were new, and 9,406 putatively causal variants. New associations included Japanese-specific coding, splicing and noncoding variants, exemplified by a damaging missense variant rs730881101 in TNNT2 associated with lower heart function and increased risk for heart failure (P = 1.4 × 10−15 and odds ratio = 4.5, 95% confidence interval = 3.1–6.5). Putative causal noncoding variants were supported by state-of-art in silico functional assays and had comparable effect sizes to coding variants. A plausible example of new mechanisms of causal variants is an enrichment of causal variants in 3′ untranslated regions (UTRs), including the Japanese-specific rs13306436 in IL6 associated with pro-inflammatory traits and protection against tuberculosis. We experimentally showed that transcripts with rs13306436 are resistant to mRNA degradation by regnase-1, an RNA-binding protein. Our study provides a list of fine-mapped causal variants to be tested for functionality and underscores the importance of sequencing, genotyping and association efforts in diverse populations.
Genome-wide association studies (GWAS) have identified thousands of loci associated with diseases and traits and have contributed to our molecular understanding of human phenotypes1,2,3,4,5,6,7,8,9. However, for most of these loci, we still do not fully understand the causal mechanisms of the associations. This is partly because of insufficient resolution of associations and limited population sources of genetic associations. Non-European large-scale association studies with sufficient resolution of variants would expand the causal mechanisms implicated by population-specific associations and variants. Additionally, the limited availability of sensitive fine-mapping strategies has hindered our understanding of causal variants10,11. Furthermore, a substantial fraction of the lead variants and their linked variants exist in noncoding regions, where functional interpretation is still challenging. Enrichment of causal variants in functional annotations would provide clues about the underlying mechanisms12.