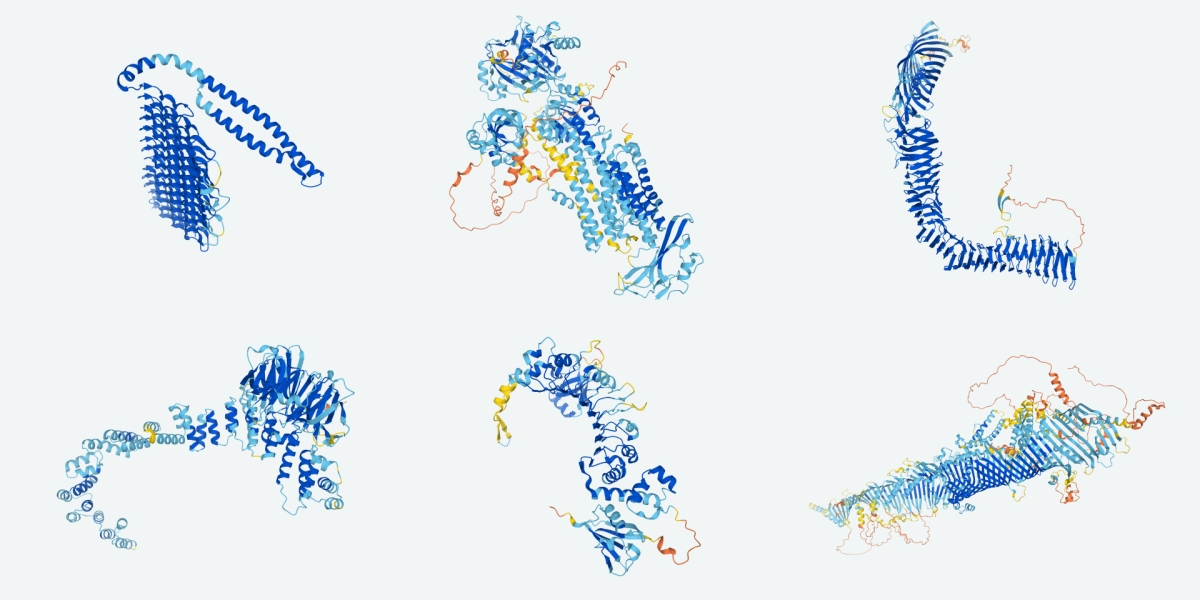
New public database of AI-predicted protein structures could transform biology
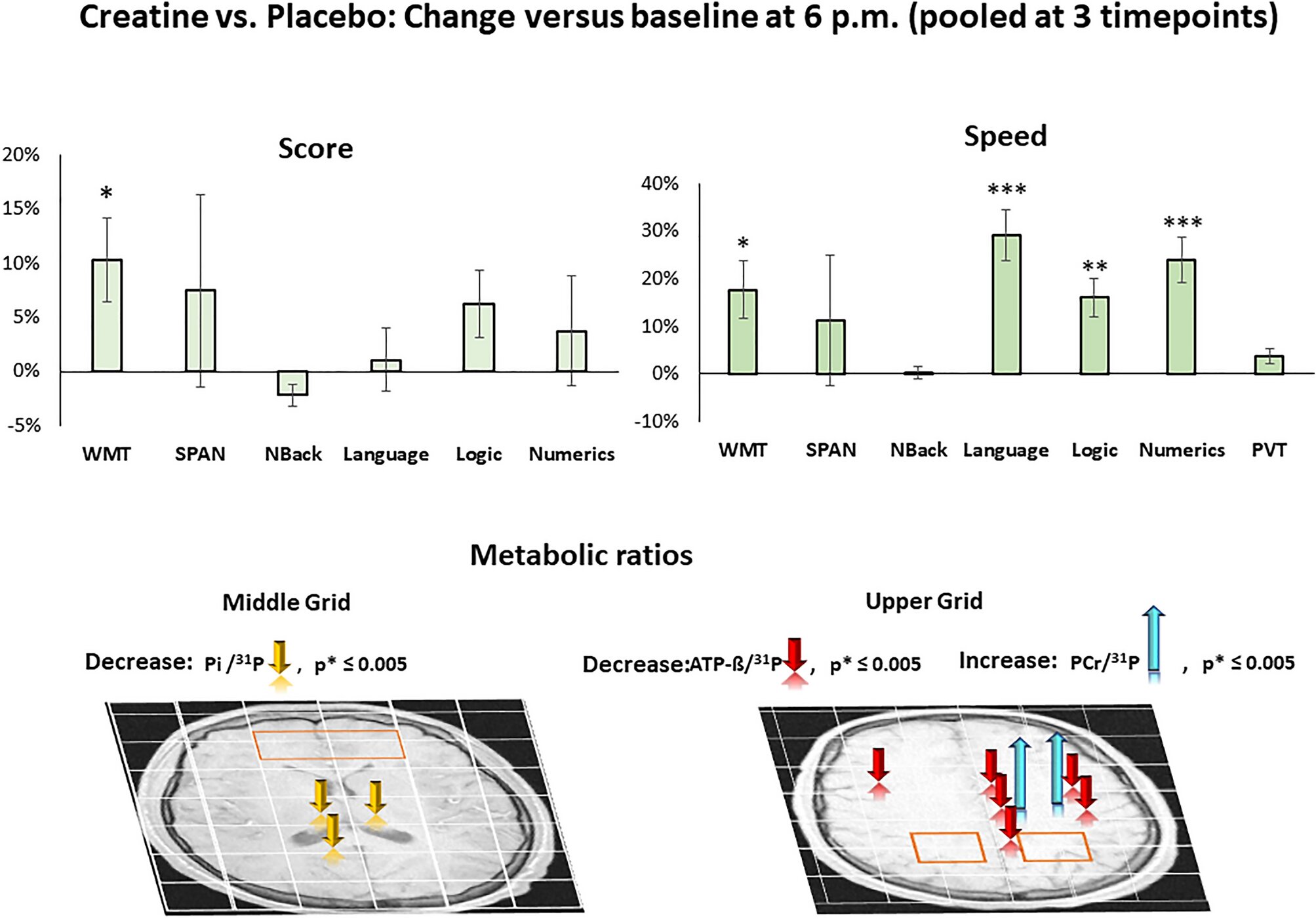
Last week, two groups unveiled the culmination of years of work by computer scientists, biologists, and physicists: advanced modeling programs that can predict the precise 3D atomic structures of proteins and some molecular complexes. And now, the biggest payoff of that work has arrived. One of those teams reports today it has used its newly minted artificial intelligence (AI) programs to solve the structures of 350,000 proteins from humans and 20 model organisms, such as Escherichia coli bacteria, yeast, and fruit flies, all mainstays of biological research. In the coming months, the group says it plans to expand its list of modeled proteins to cover all cataloged proteins, some 100 million molecules.
“It’s pretty overwhelming,” says John Moult, a protein folding expert at the University of Maryland, Shady Grove, who runs a biennial competition called the Critical Assessment of protein Structure Prediction (CASP). Moult says structural biologists have dreamed for decades that accurate computer models would one day augment extremely precise protein shapes derived from experimental methods such as x-ray crystallography. “I never thought the dream would come true,” Moult says.
The computer model, called AlphaFold, is the work of researchers at DeepMind, a U.K. AI company owned by Alphabet, the parent company of Google. In fall of 2020, AlphaFold swept the CASP competition, tallying a median accuracy score of 92.4 out of 100, well ahead of the next closest competitor. But because DeepMind researchers didn’t reveal the details of how they mapped protein shapes theoretically, specifically AlphaFold’s underlying computer code, other teams were left frustrated, unable to build on the progress. That began to change last week. On 15 July, researchers led by Minkyung Baek and David Baker at the University of Washington, Seattle, reported online in Science that they had created a highly accurate protein structure prediction program called RoseTTAFold, which they released publicly. The same day, Nature rushed out details of AlphaFold in a paper by DeepMind researchers led by Demis Hassabis and John Jumper.








/cdn.vox-cdn.com/uploads/chorus_asset/file/24801728/Screenshot_2023_07_21_at_1.45.12_PM.jpeg)